July 30, 2015 report
Closer look offers clues on how DNA strands are separated during replication
(Phys.org)—A team of researchers working in China has used newly developed cryo-electron technology to get a closer look at the mechanism involved in separating DNA strands necessary for natural replication of DNA. In their paper published in the journal Nature, the team describes their study, what they were able to observe, and their theory on how the separation actually takes place. Mathew Bochman with Indiana University and Anthony Schwacha with the University of Pittsburgh offer a News & Views piece on the work done by the team in the same journal issue.
In order for DNA to be replicated naturally, the two strands that make up its double helix must be separated—how that process occurs has been the subject of much debate. In this new effort, the researchers were able to get the closest look ever at the components involved allowing them to offer a theory on the process.
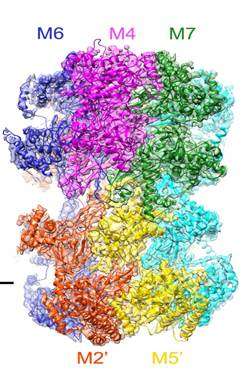
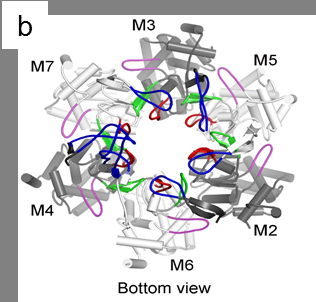
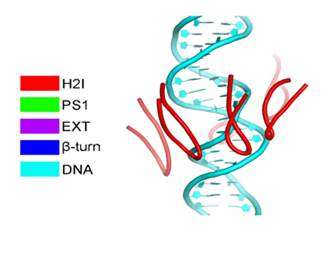
Under the new microscope, the researchers were able to see the structure of the helicase enzyme, Mcm2–7, which prior research had suggested was a major player in separating DNA strands (by somehow binding to one strand or another). The structures were doughnut shaped hexamers, two of which came together to form a single unit offset and tilted at a 14° degree angle to one another. The DNA strands would pass through both holes, via a central channel, the team noted, providing the basis, they believe for separating the two strands. Because the researchers were not able to watch as the helix was split, they were left to make an educated guess as to how the hexamers did their work. They believe that melting occurs rather than unwinding and that it occurs due to the angle between the hexamers—the narrowness of the channel combined with the angle causes, they believe, a kink to occur in the DNA strands, which they believe initiates melting of the bonds between the strands, allowing them to separate.
As for how the melting itself works, Bochman and Schwacha note that prior work has shown that an SV40 antigen has been seen to serve as a pump—they believe the hexamers might work in a similar way, noting also that the unique alignment of the hexamers allows for two exit channels—one for each of the separated strands.
More information: Structure of the eukaryotic MCM complex at 3.8 Å, Nature (2015) DOI: 10.1038/nature14685
Abstract
DNA replication in eukaryotes is strictly regulated by several mechanisms. A central step in this replication is the assembly of the heterohexameric minichromosome maintenance (MCM2–7) helicase complex at replication origins during G1 phase as an inactive double hexamer. Here, using cryo-electron microscopy, we report a near-atomic structure of the MCM2–7 double hexamer purified from yeast G1 chromatin. Our structure shows that two single hexamers, arranged in a tilted and twisted fashion through interdigitated amino-terminal domain interactions, form a kinked central channel. Four constricted rings consisting of conserved interior β-hairpins from the two single hexamers create a narrow passageway that tightly fits duplex DNA. This narrow passageway, reinforced by the offset of the two single hexamers at the double hexamer interface, is flanked by two pairs of gate-forming subunits, MCM2 and MCM5. These unusual features of the twisted and tilted single hexamers suggest a concerted mechanism for the melting of origin DNA that requires structural deformation of the intervening DNA.
Journal information: Nature
© 2015 Phys.org