September 23, 2015 report
The hidden evolutionary relationship between pigs and primates revealed by genome-wide study of transposable elements
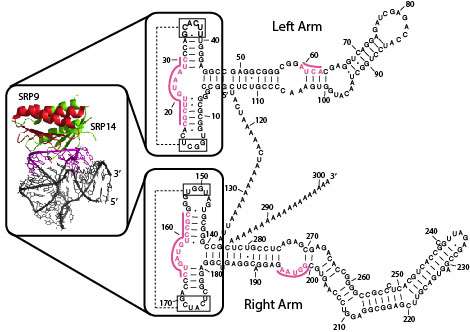
(Phys.org)—In the past, geneticists focused primarily on the evolution of genes in order to trace the relationships between species. More recently, genetic elements called SINEs (short interspersed elements) have emerged as a much better way to trace mammalian phylogeny, at least in the time since its massive radiation some 60 million years ago. That's because the prolific SINE family evolved differently in every lineage to become widespread throughout the entire genome of each. But SINEs are more than just highly mobile markers, they have specific functions—functions which researchers are now decoding to understand not just how, but why they move about like they do.
In humans, the most familiar and abundant SINE is the Alu transposable element. Originally derived from a small cytoplasmic signal recognition conglomerate known as the 7SL RNA, Alu inserts have since propagated themselves to generate an extended family over a million strong representing 11% of our entire genome. With the exception of the Alu inserts in the primate superfamily (and one seemingly anomalous occurrence of the 'B1' SINE the rat) all other SINEs were believed to have been derived from tRNAs rather than the 7SL RNA.
A recent paper published in the bioRxiv now suggests that another species—the pig—has a unique family of SINEs whose evolution has closely paralleled ours. This collaboration between researchers from China, and Firefly Bioworks Inc. here in the US, reports that the swine SINE known as PRE-1 (for porcine repeat element), also likely derives from 7SL RNA. This work potentially pushes back the divergence time of 7SL RNA products to 80-100 million years ago—a re-adjustment that would presumably ground the 7SL RNA diversification or hybridization events to a place before the so-called boreoeutherians diversified into Laurasiatheria and Euarchontoglires.

Who and exactly what were these generously-named beasts you might be asking yourself? The Laurasiatheria are the placental mammals believed to have hailed from the northern supercontinent of Laurasia after it split from Gondwana when Pangaea broke up. Their sister group, the Euarchontoglires, are the Supraprimates. These consensus classifications were made using the larger family of retrotransposons of which SINEs, and longer related LINEs, are themselves members of.
In the pig genetics business, the preferred classification term for the family is 'suidae'. Suidae PRE elements have been known since their original discovery back in 1987. The researchers identified the PRE1 element as a polymorphic insertion in the 5'-flanking region (about 686~985 bp upstream from the transcription initiator ATG codon) of the insulin growth factor binding protein (IGFBP7). Postulating that this 300 base pair insert sequence might be related to Alu inserts, they compared it with representative primate Alu sequences selected from the AF-1 database (software.iiar.res.in/af1/index.html). Incidentally, we should note that pig geneticists don't have a monopoly on the term PRF-1. In related areas of pursuit, like drosophila genetics for example, a search for 'PRE-1' might be just as likely to return 'photoreceptor regulatory element 1'.
The researchers were able to identify additional PRE-1 elements using a 'BLAST' search of similar sequences. They also were able to predict the secondary assembled structure of PRE-1 RNA using RNAstructure Webservers ( rna.urmc.rochester.edu/RNAstru … edict1/Predict1.html ). The upshot of all this work, in the author's own words, is that the 'genomic performance of PRE-1 in terms of 7SL RNA-derived SINEs seemed convincing enough to classify the suidae into a family mainly inhabited by primates'.
It has not escaped our attention that many readers of physorg who may have availed themselves of our previous coverage of the human hybrid origins theory, and a later follow up report, might come to premature conclusions here. With that in mind, I talked the creator of the hybrid origins theory, Eugene McCarthy, to get his take on this new research. He had this to say:
"People have been congratulating me on this SINE study as if it somehow proved the hybrid theory of human origins. That's nice of them, but it's just one run in a nine-inning game. True, it does show that pigs are more closely related to primates than has generally been thought, which in turn suggests that a hybrid cross between pig and chimpanzee is more feasible than many have supposed. But to establish whether we're actually descendants of an ancient cross between pig and chimpanzee, will require a detailed search of the human genome, not just a study of SINEs. I've explained the relevant issues in a recent rebuttal of some of the most common criticisms of the hybrid theory. It's not as simple as some people like to think."
More information: biorxiv.org/content/biorxiv/ea … 8/31/025791.full.pdf
© 2015 Phys.org