Machine learning applications in drug repurposing
As of January 27, the global tally of SARS-CoV-2 infections has surpassed 100 million, with fatalities reaching 2 million. Remarkably, approximately 30 million confirmed cases remain untreated with specific antiviral drugs.
Addressing this, two primary strategies have emerged to identify effective treatments: de novo drug design and drug repurposing. De novo drug design involves the discovery of novel drugs or drug combinations, starting with an analysis of the receptor protein's structure. In contrast, drug repurposing seeks to identify new therapeutic applications for existing drugs or drug combinations.
De novo drug design
The landscape of de novo drug discovery faces formidable challenges, including prolonged research and development cycles, substantial costs, and a relatively low experimental success rate. In recent years, there has been a noticeable delay in the pace of drug research and development within the pharmaceutical industry.
However, integrating machine learning techniques in mining compound properties and activities presents a promising avenue to mitigate these challenges, potentially reducing both time and costs efficiently.
A pivotal aspect of this approach is the representation of drug compounds through molecular fingerprints. These fingerprints, which can be static or dynamically generated, are crucial in characterizing compound properties. Dynamic fingerprints, in particular, are generated automatically through the training process using machine learning algorithms.
Notably, representation learning, which leverages neural network-based methods, allows for the direct training and embedding of compound features. This methodology offers a more nuanced and potentially more effective means of understanding and predicting compound behaviors and interactions, thereby streamlining the drug discovery process.
Drug repurposing
The strategy of de novo drug discovery plays a pivotal role in enriching the pharmaceutical landscape. However, this approach often incurs prohibitive costs and extended timelines. In contrast, leveraging existing drugs with well-understood mechanisms of action and pharmacokinetics offers a foundational knowledge base for novel applications.
This approach, known as drug repurposing, capitalizes on the established safety and efficacy profiles of existing drugs, thereby circumventing the need to start from scratch.
Drug repurposing has emerged as a viable and increasingly popular strategy, garnering attention from governments and pharmaceutical companies alike. Its ability to deliver time and cost efficiencies in drug development is noteworthy. Furthermore, the integration of AI technology in this domain has further enhanced the efficiency of drug repurposing processes, optimizing both time and economic resources.
A notable advantage of drug repurposing lies in its synergy with new computational methods that enable the detection of intricate relationships among various biological entities, including genes, proteins, diseases, and drugs. This study underscores the potential of drug repurposing in identifying new therapeutic indications for existing drugs.
The onset of the COVID-19 pandemic has further accentuated the relevance of drug repurposing. Amidst the lengthy cycle associated with de novo drug design, drug repurposing has become a preferred method for rapidly identifying therapeutic drugs or potential drug combinations. Drugs such as chloroquine and remdesivir have been repurposed to evaluate their therapeutic effects against COVID-19.
Compared to traditional drug discovery approaches, drug repurposing offers superior efficiency and cost-effectiveness. Moreover, it presents distinct advantages in addressing pandemic scenarios like COVID-19, where timely and cost-effective solutions are imperative.
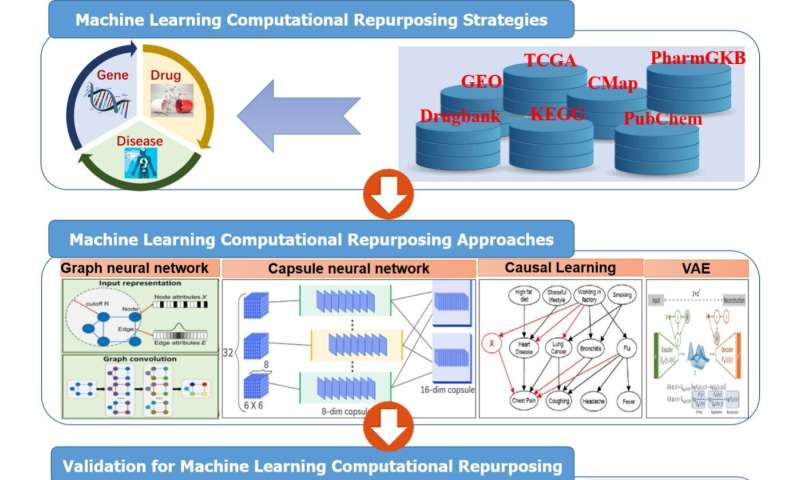
Machine learning for drug repurposing
Machine learning methodologies are instrumental in advancing the field of drug repurposing. Traditional machine learning techniques, including Logistic Regression, Random Forest, Support Vector Machines (SVM), K-Nearest Neighbors (KNN), and RotatE, have been pivotal in the initial stages of this domain. These methods have laid the groundwork for more advanced analytical processes in drug repurposing.
In recent decades, the emergence of deep learning technologies has revolutionized the potential for identifying and repurposing drugs. Deep learning models, notably Recurrent Neural Networks (RNN), Graph Convolutional Networks (GCN), Convolutional Neural Networks (CNN), Graph Neural Networks (GNN), Long Short-Term Memory networks (LSTM), Variational Autoencoders (VAE), and Transformer-based methods, have demonstrated remarkable efficacy in this area.
The superior capability of deep learning models to process and interpret complex datasets allows for more effective identification of repurposable drugs.
A significant advantage of deep learning over traditional methods lies in its ability to extract more intricate and informative features from molecular data. These models excel at mapping molecular structures to potential application spaces, providing a more nuanced understanding of how existing drugs can be repurposed. Integrating deep learning in drug repurposing represents a major leap forward in the field, offering promising avenues for pharmaceutical research and development.
The findings are published in the journal Interdisciplinary Sciences: Computational Life Sciences.
More information:
Fan Yang et al, Machine Learning Applications in Drug Repurposing, Interdisciplinary Sciences: Computational Life Sciences (2022). DOI: 10.1007/s12539-021-00487-8
Provided by Shanghai Jiao Tong University Journal Center