The detection of early-stage lung cancer by exosomal and free nucleic acids and standard screening methods
The early detection of lung cancer (LC) remains a critical challenge in oncology. This review explores the potential of exosomes (Exs) and cell-free nucleic acids (cfNAs) in the bloodstream as biomarkers for early LC detection. Exosomes, small extracellular vesicles, carry nucleic acids and proteins that reflect the molecular state of their cells of origin. This review compares the efficacy of Exs and cfNAs, emphasizing their diagnostic capabilities and limitations.
Exosomes are nano-sized vesicles secreted by various cell types, including tumor cells. They contain a range of DNA, RNA, and proteins that can influence recipient cells. Exosomal DNA (exoDNA) and RNA (exoRNA) can indicate the presence of tumors at an early stage. Liquid biopsies leveraging Exs offer a non-invasive method for detecting cancers before clinical symptoms appear, providing a potential advantage over traditional imaging techniques like CT/PET scans and MRIs.
Approximately 7% of cell-free DNA (cfDNA) in the bloodstream is not encapsulated within Exs. These include DNA fragments from virtosomes and mitochondrial DNA, among others. Tumor-derived virtosomes can transfer oncogenic DNA to healthy cells, potentially transforming them into tumor cells. This highlights the importance of considering non-exosomal cfDNA in cancer diagnostics. Bacterial and viral DNAs also contribute to the cfDNA pool, some of which may be related to tumor viruses, serving as biomarkers for cancer detection.
The distribution of RNA between exosomal and non-exosomal fractions is less well-studied. Methods to extract and measure miRNA from plasma and other bodily fluids exist, but the proportion of total versus exosomal RNA remains unclear. Understanding this distribution is essential for optimizing RNA-based diagnostics.
Exosomes are rapidly cleared from the bloodstream by organs such as the liver, spleen, kidneys, and lungs. Studies using fluorescently labeled Exs demonstrate their quick uptake by these organs, with a half-life in circulation of about 2 minutes. This rapid clearance poses a challenge for the detection of exosomal biomarkers, as their short-lived presence in blood may hinder timely diagnosis.
CfDNA is also swiftly removed from circulation, primarily by the liver's lysosomal system. Single-stranded cfDNA (sscfDNA) and double-stranded cfDNA (dscfDNA) exhibit different uptake rates, with sscfDNA being more readily processed and excreted. This efficient removal system complicates the use of cfDNA as a diagnostic tool, as its rapid clearance could lead to false negatives in cancer detection.
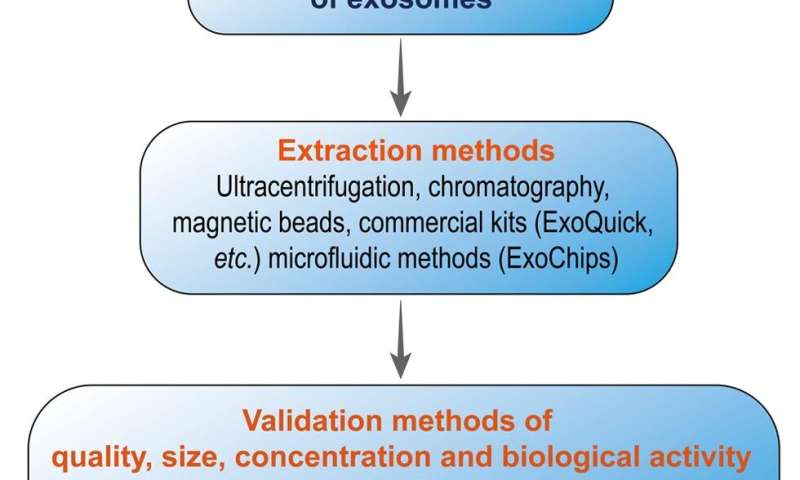
The isolation of Exs and nucleic acids involves several methodologies. Differential ultracentrifugation, PEG-based precipitation, size exclusion chromatography, and surface protein capture are common techniques. Each method has its advantages and limitations. For example, differential ultracentrifugation is widely used but is time-consuming and may affect the quality of Exs. PEG-based methods are simpler but may result in lower purity.
Exs and cfNAs hold significant promise as biomarkers for early LC detection. While exosomal nucleic acids offer insights into the molecular characteristics of tumors, their rapid clearance from the bloodstream presents a challenge. Similarly, non-exosomal cfDNA provides valuable information but is also quickly removed. Optimizing the techniques for isolating and analyzing these biomarkers is crucial for improving early cancer diagnosis and patient outcomes.
By addressing these challenges and refining diagnostic methods, the potential for early, non-invasive detection of LC using Exs and cfNAs could be realized, paving the way for better prognosis and treatment strategies.
The paper is published in the journal Cancer Screening and Prevention.
More information:
Peter B. Gahan et al, A Comparative Review of the Detection of Early-stage Lung Cancer by Exosomal and Free Nucleic Acids and Standard Screening Methods, Cancer Screening and Prevention (2023). DOI: 10.14218/CSP.2022.00021
Provided by Xia & He Publishing Inc.