Aqueous protons can be found in protein X-ray structures
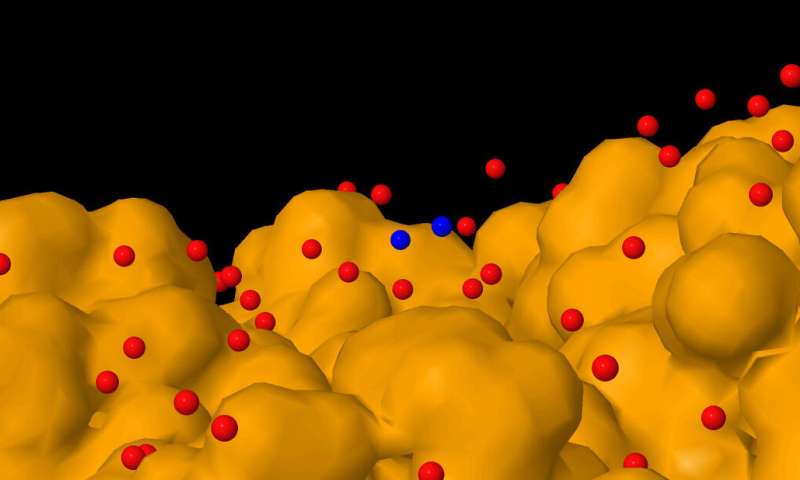
It is possible to determine the presence and location of aqueous proton species at the surface and in the cavities of proteins, as a recently published study shows. This finding goes against conventional view and sheds light on how protons (and hydroxides) participate in a huge number of processes.
The aqueous proton is one of the most ubiquitous and important species in chemistry. The processes in which this chemical species (and the related aqueous hydroxide) participates are practically innumerable. It probably represents the most common enzyme substrate, as it is involved in an impressive number of enzymatic reactions. However, until today, the structural evidence of aqueous protons in proteins, and macromolecules in general, were frankly scarce.
The most popular technique for determining the structure of macromolecules is X-ray diffraction: Almost 90 percent of the structures currently available in the Protein Data Bank (PDB) have been obtained using this approach. A significant number of these structures has been resolved at atomic resolution. Since X-ray diffraction is due to the electrons in the crystallized sample, protons are practically invisible to this technique because they are electron-poor species. A small number of protons belonging to proteins is visible in very high resolution structures, but in general, the protons of the solvent (i.e., water) are not visible even in these structures. The limited structural evidence of this type has been obtained by neutron diffraction, which, being sensitive to the presence of nucleons, is also able to detect hydrogen atoms.
Protons are invisible in X-ray determined structures, but ...
Analyzing the radial distribution function of water in a large dataset of high-resolution X-ray structures, Palese found some intriguing features in the oxygen-oxygen distances of the protein-bound water molecules. The highest peak in the radial distribution function of water oxygen atoms was evidently attributable to the presence of hydrogen bonds. The shape of this peak suggested the presence of two populations, one similar to liquid water, and another akin to ice. But the most astounding finding was the presence of an anomalous peak at an oxygen-oxygen distance shorter than expected, which suggested the presence of water organized to form pairs, or clusters, of molecules positioned at short distance from each other in the water network around the proteins.
After eliminating all plausible artifacts, a probability excess at this anomalously low distance still remained. The simplest explanation for these clusters of short-distance water molecules was that they were involved in the binding of an excess of protons. Oxygen-oxygen distances below 2.70 Å can be related to the presence of a H+ bridge between two neighboring water molecules, as in the Zundel ion. Similar short distances (or those even shorter) are expected in the case of aqueous hydroxide.
The position of these short-distance water molecules was analyzed in model proteins that were chosen for an in-depth analysis because they are involved in well-known proton transfer processes. These proteins were cytochrome c oxidase, a mitochondrial proton pump, and carbonic anhydrase, a ubiquitous enzyme involved in in acid-base homeostasis. The intriguing finding was that these short-distance water molecules were mainly positioned at the protein surface, and particularly near the entry mouth of proton channels and near antenna regions (i.e., regions at the protein surface able to irradiate and/or receive protons).
Why can we locate an extremely diffusive chemical species?
But how to explain the fact that a species with high diffusivity like the aqueous proton could be seen in a particular region of the protein? Palese proposes that the aqueous proton "lives" in the network of water molecules, and even if mobile, it can be located preferentially in some regions (such as, for example, in the antenna regions on the protein surface). X-ray exposure time in a diffraction experiment is typically on the order of seconds; in the case of mobile protons, we must also expect a shortening of the oxygen-oxygen distance of the water molecules that bind them preferentially. In reality, we do not actually see the proton, but observe the effects of its permanence in some structures.
The study not only showed that it is possible to "visualize" the aqueous protons in proteins, but also provided details on their localization that are in perfect agreement with a large body of data and of theories. Moreover, this study suggested that the dissociated forms of water could be considered among the most important counter-ions of proteins.
This story is part of Science X Dialog, where researchers can report findings from their published research articles. Visit this page for information about ScienceX Dialog and how to participate.
More information:
Luigi Leonardo Palese. Oxygen-oxygen distances in protein-bound crystallographic water suggest the presence of protonated clusters, Biochimica et Biophysica Acta (BBA) - General Subjects (2019). DOI: 10.1016/j.bbagen.2019.129480