Discovery of a conserved anti-SARS-CoV-potential therapeutic target in the heart of human and bat spike proteins
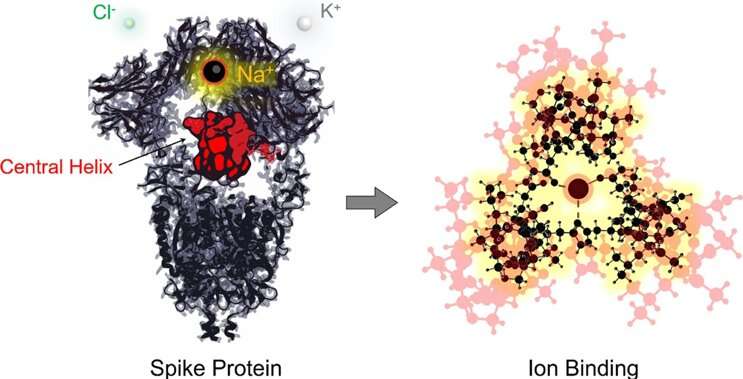
The spike glycoprotein of the human severe acute respiratory syndrome coronaviruses (hSARS-CoVs) represents an attractive target for the design and development of biopharmaceuticals as well as small molecule drugs, desirable in terms of either preventing infection or effectively treating disease progression. Despite the effort spent in the development and large-scale production of vaccines to manage the actual COVID-19 pandemic, surges in SARS-CoV-related hospitalizations still represent a major risk for several countries. Nonetheless, first-line antiviral drugs are missing in clinical practice, especially for severely affected patients who may need a specific pharmacological treatment.
The spike protein funnel
As computational chemists, we decided to inspect available human SARS-CoV-2 spike-protein structures as recently solved by crystallographers, searching for any possible interesting site other than the ACE2-receptor binding domain (generally studied by scientists for the design of vaccines and spike-directed antibodies).
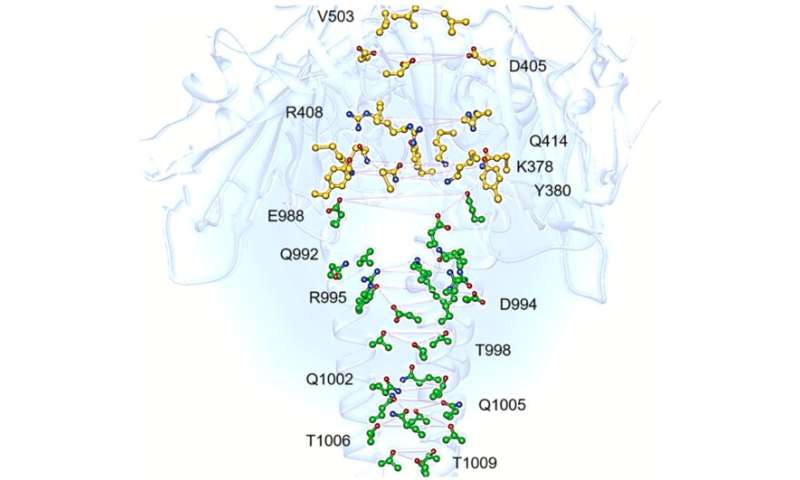
We were impressed by the elegant funnel shape assumed by the central helix domain in the state preceding fusion with the human cell. Our immediate question was "How important is this central motif?" After looking at the alpha-helix hydrogen bond path leading to the bottom of the protein funnel, in representative open and closed structures, it was clear that something structurally important was happening in that region. Both cryo-EM data and chemical intuition led us finally to the idea that ions could be bound in the latter.
The ion binding site of SARS-CoV spike glycoproteins
Interestingly, we found sequence conservation of the central helix residues to be 100% in human and bat SARS-like coronaviruses, suggesting an evolutionary role of this site. SARS-CoV-1 spike-protein structures, corresponding to the previous SARS-CoV epidemics of 2003, were also available, and we now know that the same protein can effectively bind chloride ions after fusion with the cell membrane (see Duquerroy et al, 2005). So what about the pre-infection state? Is it also able to bind ions? Unfortunately, cryo-EM structures do not offer enough resolution to ion and water molecule visualization, but our DFT quantum-chemical calculations, recently published in Computational and Theoretical Chemistry, overcome this limit and demonstrate that physiological electrolytes (sodium, potassium and chloride ions) can be easily bound by that central helix site, shared between SARS-like CoV spike proteins, especially sodium cation (Figure 2).
Conclusions and perspectives
The high conservation of the identified binding site indicates that it should have a substantial role in the spike-protein functions of any SARS-like coronavirus. Specifically, this aspect would be in line with the sequestration of electrolytes from the physiological solution, as probably a key result of the evolutionary adaptation to the mammalian cells' environment. Conservation of the site and also its general binding features suggest that small molecules may be specifically designed in order to interfere with the helix domain structure of either current or future SARS-CoVs, paving the way to new potential antiviral drugs for the clinical practice.
This story is part of Science X Dialog, where researchers can report findings from their published research articles. Visit this page for information about ScienceX Dialog and how to participate.
More information: Stéphane Duquerroy et al, Central ions and lateral asparagine/glutamine zippers stabilize the post-fusion hairpin conformation of the SARS coronavirus spike glycoprotein, Virology (2005). DOI: 10.1016/j.virol.2005.02.022
Enrico Margiotta et al, SARS-CoV spike proteins can compete for electrolytes in physiological fluids according to structure-based quantum-chemical calculations, Computational and Theoretical Chemistry (2021). DOI: 10.1016/j.comptc.2021.113392
Enrico Margiotta is an Italian computational chemist. He was born in 1990 in Enna (Sicily, Italy). He was awarded a joint-degree PhD in molecular modeling by the University of Padua (Italy) and the Vrije Universiteit Amsterdam in 2020. Enrico specializes in drug design and quantum DFT methods for medicinal chemistry, and is currently working as a research scientist at the Broad Institute of MIT and Harvard in Boston.